Areas of Research
Take a look at what we’re working on…
Current Projects
In Monir lab, we employ lab experiments, field-research and bioinformatic approaches to pursue several major research directions:
- Given that giant viruses are highly diverse and extremely common in both mairne and freshwater ecosystems, it is important to determine the ecological impacts of these viruses. A major focus of the lab is to determine the role of giant viruses within the microbial community and how interactions of these viruses with diverse unicellular eukaryotes drive their population dynamics and contribution in the food web. To address these questions, we integrate multi-omics datasets from time-series studies in coastal and open ocean environments.
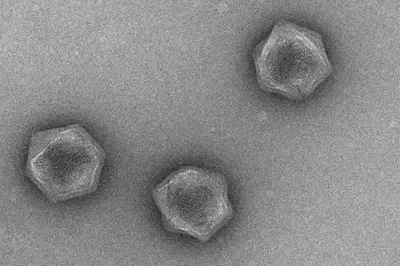
2. Deciphering the molecular underpinnings of ‘virocell’ metabolism: A ‘virocell’ is a conceptual framework that emphasizes the fact that lytic viral infection remodels the physiology of a host cell. Viral lysis of numerous microbial taxa can modulate the nutrient flux within the marine microbiome and also contributes to the efficiency of the biological carbon pump. Understanding the molecular underpinnings of virocell metabolism is therefore crucial for evaluating the contribution of viruses to the global. In eukaryotic cells, the presence of large genomes and distinct organelles result in a highly complex physiological landscape that viruses must rewire for successful propagation. Additionally, giant viruses often encode a large number of metabolic genes. However, there is only limited information on the precise role of these genes in manipulating the nutrient flux, energy and carbon metabolism of infected host cells. A key focus of our research is to investigate the molecular remodeling of the virocell, and the role of giant virus-encoded metabolic genes - leveraging the existing host-virus systems already available in culture.
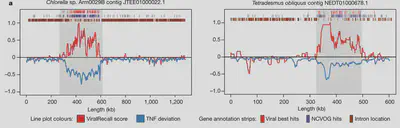
3. Assessing the impact of endogenous giant viruses on the physiology and genome evolution of their eukaryotic hosts: Endogenous viral elements (EVEs)—viral sequences that integrate into host genomes and eventually become heritable as host alleles—can spur host genome rearrangement, functional innovations, and mediate horizontal gene transfer (HGT). In our recent work, we have identified widespread endogenization of NCLDV genomes in diverse green algae. These Giant Endogenous Viral Elements (GEVEs) are several hundred kilobases long and contribute up to 10% of the total genes in some of the chlorophyte genomes in which they are integrated. In future work, we plan to develop a foundational understanding of the role endogenous NCLDVs play in the genome evolution, physiology and phenotypic diversity of protists